Notre groupe étudie les fonctions biologiques de l'hôte mobilisées lors de la réplication et la pathogenèse des virus humains et la réponse à ces infections.
Nous explorons la relation entre le VIH-1 (virus de l’immunodéficience humaine), agent causal du SIDA, et la cellule hôte afin de comprendre les mécanismes moléculaires qui contrôlent la réplication et la propagation virale et d’identifier les stratégies potentielles de contrôle ou d'éradication de ce virus.
- Le laboratoire décrypte les mécanismes de latence du VIH qui conduisent à la persistance de cellules hébergeant des virus compétents pour la réplication mais transcriptionnellement silencieux. Ces réservoirs, établis tôt au cours de l'infection, peuvent persister pendant une période prolongée. Si le traitement est interrompu, le rebond viral se produit rapidement.
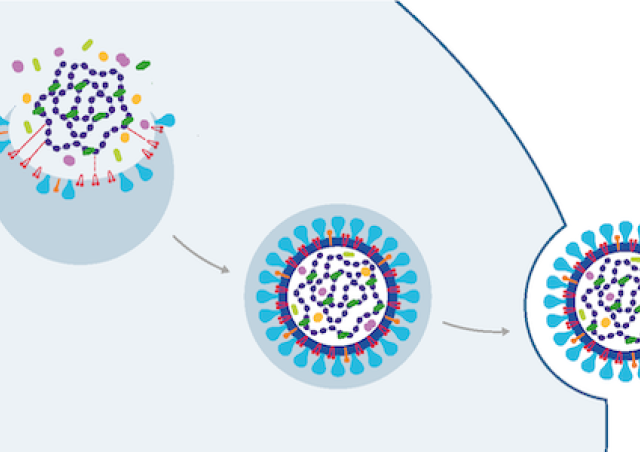
- Nous explorons également la base moléculaire de l'expression du VIH-1 tant au niveau transcriptionnel que post-transcriptionnel. Le transcriptome du VIH-1 est complexe et doit être finement régulé pour permettre la morphogénèse de particules virales infectieuses.
- L’équipe étudie aussi comment le VIH-1 contrecarre les facteurs de restrictions, protéines cellulaires possédant des activités antivirales directes. Ces facteurs constituent une facette de la réponse immunitaire innée et contrôlent l'infection de manière intrinsèque à la cellule. Nous étudions notamment les restrictions qui s’opposent aux virus lors de l'assemblage et la libération des virions.
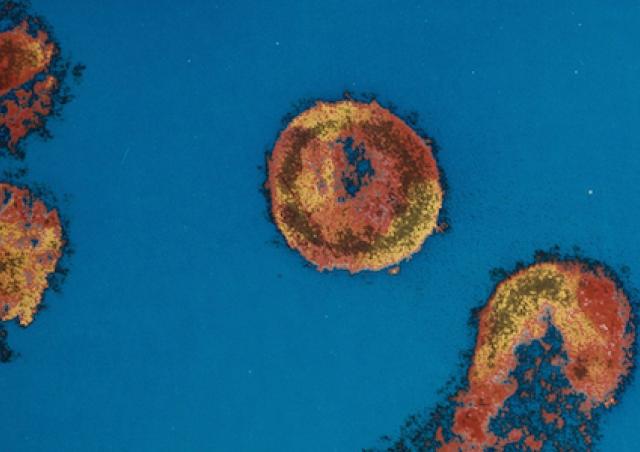
L'émergence du coronavirus SARS-CoV-2 et sa rapide propagation au niveau mondial nous ont récemment conduit à explorer les interactions hôte-virus impliquées dans la réplication de ce pathogène. Des approches protéomiques sont développées pour caractériser les facteurs cellulaires indispensables à la morphogénèse de SARS-CoV2 infectieux.
Pour répondre à ces objectifs, nous utilisons des techniques de génétique, de biologie cellulaire et moléculaire, d’imagerie et de virologie.