Members of the project
Research projects
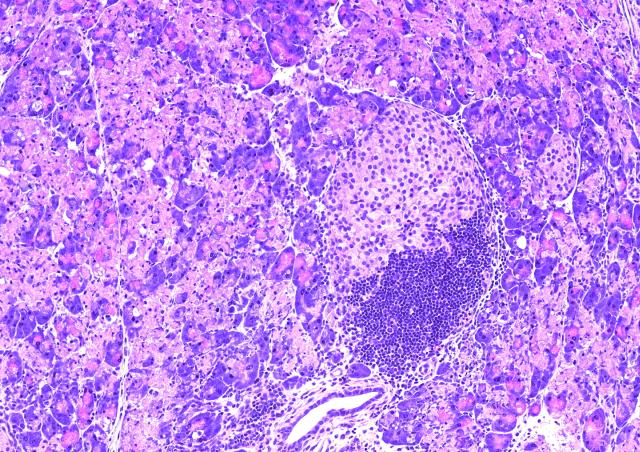
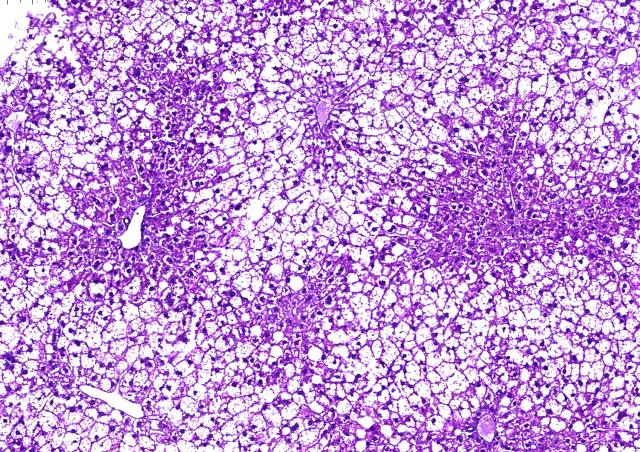
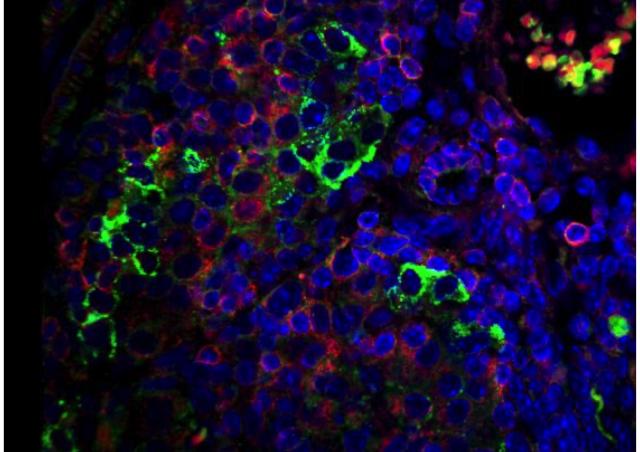
In order to identify and characterize genes involved in the diversity of diabetes, we have assembled a unique set of cohorts of patients and families, including « typical » T1D cohorts, French and international, and cohorts enriched in monogenic forms of diabetes according to various selection criteria. We use clinical, genetics and bioinformatics approach, as well as functional studies depending on the nature of identified genes. Our studies of atypical forms of diabetes resulted in the identification of several novel genes responsible for monogenic forms of diabetes, including EIF2AK3, GLIS3, DUT et ONECUT1/HNF6, and in the detailed clinical characterization of novel monogenic entities. Our studies take into account familial and populationnal components, resulting in increased power. Some of these genes are shared between multiple diabetes forms, such as GLIS3 (monogenic diabetes and multifactorial T1D and T2D) and ONECUT1 (from monogenic recessive neonatal syndromic diabetes to multifactorial T2D). The identification and study of these genes therefore provide a better understanding of mechanisms involved in diabetes. Our studies also open new propects towards precision medicine, adjusted to individual genetic risks.

Selected publications
1. Delépine M, Nicolino M, Barrett T, Golamaully M, Lathrop M, Julier C. EIF2AK3, encoding translation initiation factor 2- kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nature Genetics 25: 406-409, 2000
2. Senée V, Chelala C, Duchatelet S, Feng D, Blanc H, Cossec JC, Charon C, Nicolino M, Boileau P, Cavener DR, Bougnères P, Taha D, Julier C. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat Genet 38: 682-687, 2006
3. Zalloua P, Azar S, Delepine M, Makhoul N, Blanc H, Sanyoura M, Lavergne A, Stankov K, Lemainque A, Baz P, Julier C. WFS1 mutations are frequent monogenic causes of juvenile-onset diabetes mellitus in Lebanon. Hum Mol Genet 17:4012-4021, 2008
4. Barrett JC, Clayton D, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS, and The Type 1 Diabetes Genetics Consortium. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 41, 703–707, 2009
5. Abdulkarim B*; Nicolino M*, Igoillo-Esteve M, Daures M, Romero S, Philippi A, Senée V, Lopes M, Cunha D, Harding H, Derbois C, Bendelac N, Hattersley A, Eizirik D, Ron D, Cnop M*, Julier C*. A missense mutation in PPP1R15B causes a syndrome including diabetes, short stature and microcephaly. Diabetes 64:3951-3962, 2015
6. Dos Santos RS*, Daures M*, Philippi A*, Romero S, Marselli L, Marchetti P, Senée V, Bacq D, Besse C, Baz B, Marroquí L, Ivanoff S, Masliah-Planchon J, Nicolino M, Soulier J, Socié G, Eizirik DL*, Gautier J-F*, Julier C*. dUTPase (DUT) is Mutated in a Novel Monogenic Syndrome with Diabetes and Bone Marrow Failure. Diabetes 66:1086-1096, 2017
7. Lenfant C*, Baz P*, Degavre A*, Philippi A, Senée V, Vandiedonck C, Derbois C, Nicolino M, Zalloua P, Julier C. Juvenile-onset diabetes and congenital cataract: « double-gene » mutations mimicking a syndromic diabetes presentation. Genes 8:309, 2017
8. Lytrivi M*, Senée V*, Salpea P*, Fantuzzi F*, Philippi A, Abdulkarim B, Sawatani T, Marin-Canas S, Pachera N, Degavre A, Singh P, Derbois C, Lechner D, Ladriere L, Igoillo-Esteve M, Cosentino C, Marselli L, Deleuze JF, Marchetti P, Eizirik DL, Nicolino M, Chaussenot A, Julier C*,@, Cnop M*,@. DNAJC3 deficiency induces β-cell mitochondrial apoptosis and causes syndromic young-onset diabetes. Eur J Endocrinol. 184:459-472, 2021
9. Philippi A#, Heller S#, CostaI IG#, Senée V#, Breunig M, Zhijian L, Kwon G, Illing A, Lin Q, Hohwieler M, Degavre A, Zalloua P, Liebau S, Schuster M, Krumm J, Zhang X, Geusz R, Benthuysen JR, Wang A, Goulton K, Neubauer H, Simon E, Klein T, Wagner M, Russell R, Nair G, Besse C, Olaso R, Deleuze J-F, Kuster B, Hebrok M, Seufferlein T, Sander M, Boehm BO, Oswald F, Nicolino M*, Julier C*,@, Kleger A*,@. Mutations and variants of ONECUT1 in diabetes. Nat Medicine 27:1928-1940, 2021
10. Heller S@, Melzer MK, Azoitei N, Julier C@, Kleger A@. Human Pluripotent Stem Cells Go Diabetic: A Glimpse on Monogenic Variants. Front Endocrinol 12: 648284, 2021. Review
# Equal contributions. *Equal contributions. @co-corresponding authors