Project members
Project
Despite the central role of myofibers in muscle biology, the cellular and molecular functions of the myonuclei (nuclei of myofiber) have been largely neglected. For technical reasons, very little is known about the role of the different cell populations and myonuclei subpopulations in the control of muscle plasticity. Using innovative methods (snRNAseq and RNAscope), we project to understand how the functional heterogeneity of myofiber subdomains is specified and contributes to muscle plasticity during atrophic and hypertrophic conditions depending on mechanical load. In this context, we identify and study cells and myonuclei responsible of muscle response to mechanical stress (Axis1). We also study the influence of fibers typology in the response to hypertrophic and atrophic stimuli (axis 2).
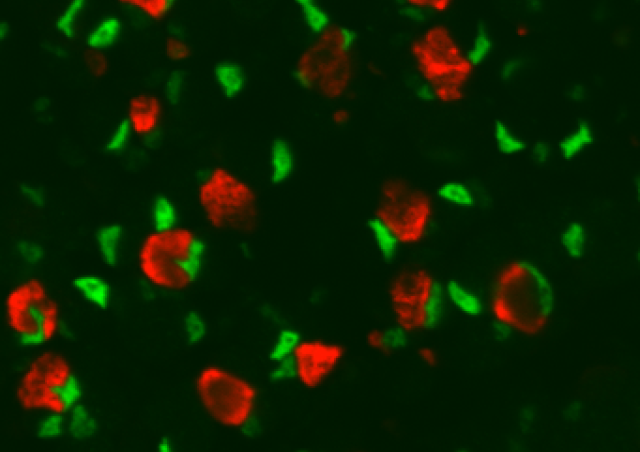
Skeletal muscle function alteration is a common feature of several diseases and is associated with an increase of patient’s mortality. Understanding of the mechanisms involved in the regulation of muscle mass, typology and metabolism is thus of clinical interest. Skeletal muscle tissue is highly heterogeneous and is composed of different cell populations such as myofibers, satellite cells, tenocyte, FAPs and macrophages. In addition, myofibers are syncytium containing more than hundred nuclei per cell divided in different subpopulations, including those of neuromuscular junction (NMJ) and myotendinous junction (MTJ) domains.
Despite the central role of myofibers in muscle biology, the cellular and molecular functions of the myonuclei (nuclei of myofiber) have been largely neglected. For technical reasons, very little is known about the role of the different cell populations and myonuclei subpopulations in the control of muscle plasticity. The snRNA-seq method (optimized in our team) permits to capture the transcriptional signature of all nuclei of the muscle tissue that will allow the identification of specific transcription profile for all myonuclei present in a muscle. Our team has also standardized the highly sensitive RNAScope FISH methodology (single RNA molecule resolution, for isolated Myofibers). This technique is complementary with snRNA-seq and permits the spatial analysis of several biomarkers. Using these innovative methods and based on strong preliminary results, we project to understand how the functional heterogeneity of myofiber subdomains is specified and contributes to muscle plasticity during atrophic and hypertrophic conditions depending on mechanical load.

Aim 1:
i) To understand the role of muscle fiber environment and heterogeneity (myonuclei subpopulation) in the adaptation to mechanical stresses.
ii) To identify molecular mechanisms involved in the crosstalk between the muscle tissue cells and/or fibers sub-domains during hypertrophy and atrophy.
Aim 2:
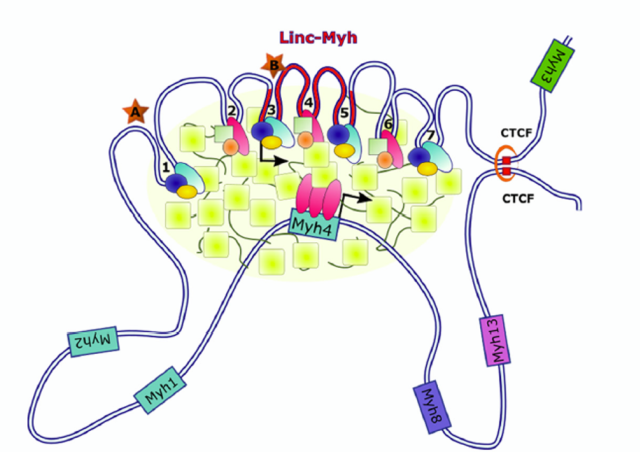
We will focus on the interplay between myofiber typology and muscle mass control. Muscle tissue can contain four different myofiber types classified by their myosin isoform expression and metabolism (Myh7 slow oxidative type I, Myh2 intermediary/slow oxidative type IIa, Myh1 intermediary/fast glycolytic IIx, Myh4 fast type IIb). Interestingly, myofibers typology varies in response to hypertrophic and atrophic stimuli. We aim to understand the mechanisms and the importance of myosin shifting in muscle plasticity. We also project to determine whether myosin isoforms expression predetermine myofibers adaptation to hypertrophy and atrophy.